
Using BaseMount¶
Introduction to BaseMount¶
What is BaseMount?
BaseMount is Illumina software that enables access to your BaseSpace storage as a Linux file system on the command line
Advantages of BaseMount:
Have access to your Projects, Runs, and App results as your local files.
Can run local apps on basespace data without downloading data to your local computer.
Can save your local storage space:
“Although BaseMount does facilitate file download, we would recommend that since BaseMount allows convenient, fast, cached access to your BaseSpace metadata and files, you may find that many operations can be carried out without the need to download locally. During our testing, we have used BaseMount to grep through fastq files, extract blocks of reads from bam files and even use IGV on the bam files directly all without downloading files locally. This can be more convenient than including a download step and saves on the overheads of local storage.” -Illumina
Official page
Getting Started with BaseMount¶
Quick Install
1 | sudo bash -c "$(curl -L https://basemount.basespace.illumina.com/install/)"
|
Manual install
Supported Operating Systems: Ubuntu, CentOS
Ubuntu 14 & 15:
1 2 3 4 | wget https://bintray.com/artifact/download/basespace/BaseSpaceFS-DEB/bsfs_1.1.631-1_amd64.deb
wget https://bintray.com/artifact/download/basespace/BaseMount-DEB/basemount_0.1.2.463-20150714_amd64.deb
sudo dpkg -i --force-confmiss bsfs_1.1.631-1_amd64.deb
sudo dpkg -i basemount_0.1.2.463-20150714_amd64.deb
|
Mounting Your BaseSpace Account
1 2 | basemount --config {config_file_prefix} {mount-point folder}
basemount --config user1 ~/BaseSpace_Mount
|
Example¶
1 2 3 | mkdir /export/NFS/Saranga/BaseSpace
mkdir /export/NFS/Maria/BaseSpace
basemount --config Maria /export/NFS/Maria/BaseSpace/
|
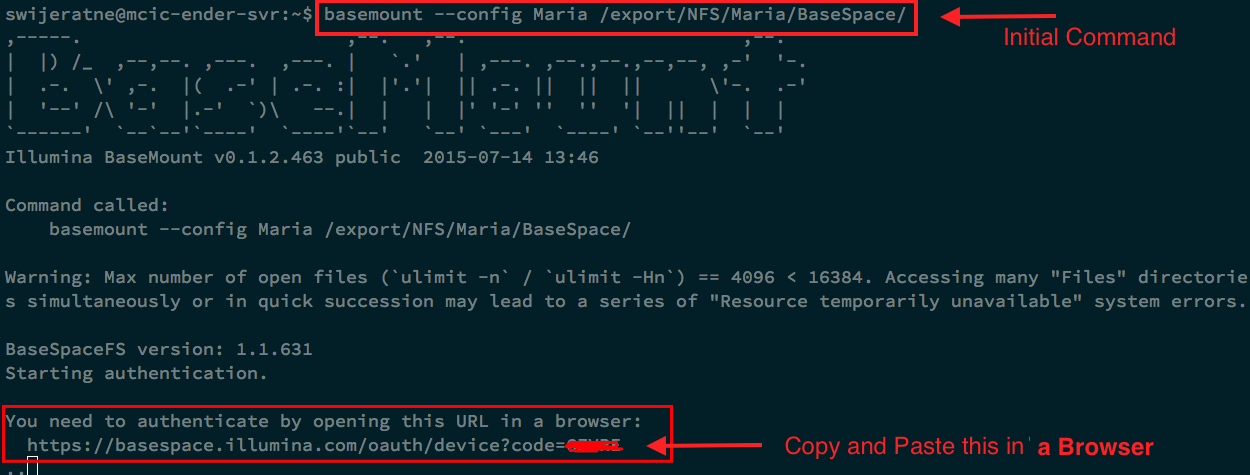
Then open your internet browser:
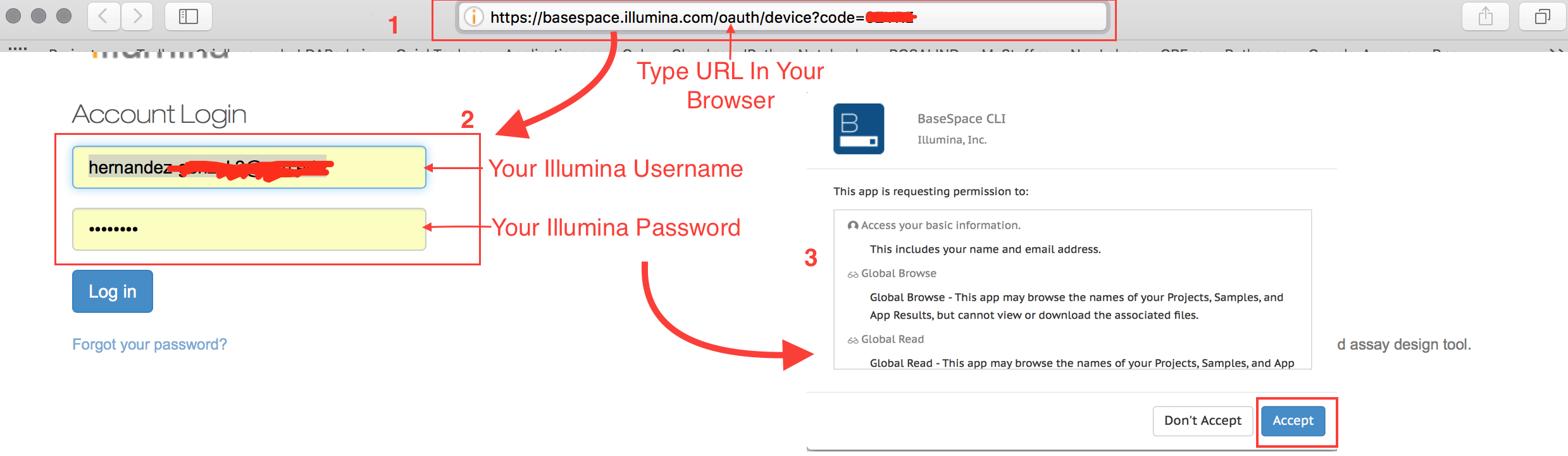
After you click “Accept”,

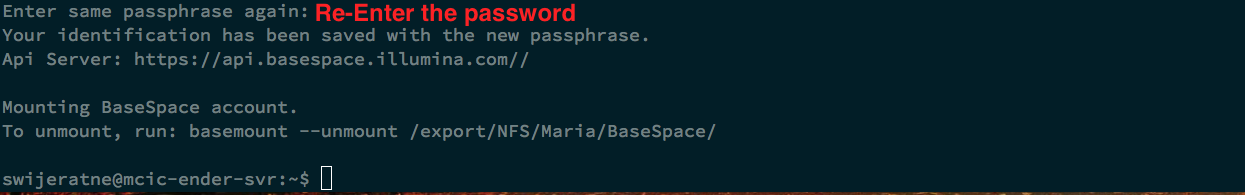
To access the folder, type:
1 2 | cd /export/NFS/Maria/BaseSpace/
ls
|
Projects Runs
To see the contents in the Projects:
1 | ls -lsh /export/NFS/Saranga/BaseSpace/Projects/HiSeq\ 2500\ -\ v4\ reagents\:\ TruSeq\ PCR\ Free\ \(4\ replicates\ of\ NA12877\)/Samples/NA12877_*/Files/
|
/export/NFS/Saranga/BaseSpace/Projects/MiSeq v3: TruSeq Targeted RNA Expression (NFkB & Cell Cycle: Human Brain-Liver-UHRR)/Samples/Brain10/Files/:
total 85M
85M -r--r--r-- 1 root root 85M Oct 5 14:09 Brain10_S22_L001_R1_001.fastq.gz
/export/NFS/Saranga/BaseSpace/Projects/MiSeq v3: TruSeq Targeted RNA Expression (NFkB & Cell Cycle: Human Brain-Liver-UHRR)/Samples/Brain11/Files/:
total 62M
62M -r--r--r-- 1 root root 62M Oct 5 14:09 Brain11_S23_L001_R1_001.fastq.gz
Basic analysis of fastq files¶
You can get basic information about your fastq files without having to download them.
For instance:
- View sequences inside Fastq files
- Get the number of reads for each
fastqfile - Get basic statistics and read length distribution
Example: View your data
1 | zcat /export/NFS/Saranga/BaseSpace/Projects/MiSeq\ v3\:\ TruSeq\ Targeted\ RNA\ Expression\ \(NFkB\ \&\ Cell\ Cycle\:\ Human\ Brain-Liver-UHRR\)/Samples/Brain1/Files/Brain1_S13_L001_R1_001.fastq.gz | head -n 4
|
@M03438:48:000000000-AGGNU:1:1101:11792:1006 1:N:0:13
NTCAATCCCCAGCAGTGGAATAAGGCCTGTTGTTCCTTGCAGTGGATCCTG
+
#88ABFFGCFEEG<FF<FDFFEGGFGGFCFGFFGGEGGGGGGFGGFGGGGG
Example: Count the number of sequences using native Linux commands
1 | zcat /export/NFS/Saranga/BaseSpace/Projects/MiSeq\ v3\:\ TruSeq\ Targeted\ RNA\ Expression\ \(NFkB\ \&\ Cell\ Cycle\:\ Human\ Brain-Liver-UHRR\)/Samples/Brain1/Files/Brain1_S13_L001_R1_001.fastq.gz | grep -c "@M03438:"
|
838876
| FILE SIZE: | 85M |
|---|---|
| TIME TAKEN: | 1.327s |
Example: Count the number of sequneces using using fastqutils .
1 | fastqutils names /export/NFS/Saranga/BaseSpace/Projects/MiSeq\ v3\:\ TruSeq\ Targeted\ RNA\ Expression\ \(NFkB\ \&\ Cell\ Cycle\:\ Human\ Brain-Liver-UHRR\)/Samples/Brain1/Files/Brain1_S13_L001_R1_001.fastq.gz | wc -l
|
838876
| FILE SIZE: | 85M |
|---|---|
| TIME TAKEN: | 23.00s |
Example: Get the Length distribution and other statistics
1 | fastqutils stats /export/NFS/Saranga/BaseSpace/Projects/MiSeq\ v3\:\ TruSeq\ Targeted\ RNA\ Expression\ \(NFkB\ \&\ Cell\ Cycle\:\ Human\ Brain-Liver-UHRR\)/Samples/Brain1/Files/Brain1_S13_L001_R1_001.fastq.gz
|
Space: basespace
Pairing: Fragmented
Quality scale: Illumina
Number of reads: 838876
Length distribution
Mean: 51.0
StdDev: 0.0
Min: 51
25 percentile: 51
Median: 51
75 percentile: 51
Max: 51
Total: 838876
Quality distribution
pos mean stdev min 25pct 50pct 75pct max count
1 33.5948650337 2.15033983948 2 34 34 34 34 838876
2 33.6285434319 2.01137931152 12 34 34 34 34 838876
3 33.6910246568 1.81306178651 11 34 34 34 34 838876
4 33.6861812711 1.84720681841 11 34 34 34 34 838876
5 33.6998292954 1.73881480815 12 34 34 34 34 838876
6 37.2788564698 2.49830369426 2 38 38 38 38 838876
7 37.3219820331 2.39576158974 2 38 38 38 38 838876
8 37.1795640834 2.72499009837 2 38 38 38 38 838876
9 37.158373824 2.76769991544 10 38 38 38 38 838876
10 37.0991517221 2.93041515545 10 38 38 38 38 838876
11 37.080357526 3.00286915879 10 38 38 38 38 838876
12 37.0752506926 2.93530617165 10 38 38 38 38 838876
13 37.1372705859 2.85104291311 10 38 38 38 38 838876
14 37.0559641711 3.00820427859 10 38 38 38 38 838876
15 37.0753877808 2.99577677236 10 38 38 38 38 838876
16 37.0950426523 2.92821324956 10 38 38 38 38 838876
17 37.204973083 2.64727500649 10 38 38 38 38 838876
18 37.1618308308 2.75627448135 10 38 38 38 38 838876
19 37.0932426247 2.92692822638 10 38 38 38 38 838876
20 37.1103548081 2.89001817524 10 38 38 38 38 838876
21 37.117659821 2.90476888945 10 38 38 38 38 838876
22 37.1280034236 2.80218109494 10 38 38 38 38 838876
23 36.9527546383 3.18334971719 9 37 38 38 38 838876
24 37.1825096915 2.70092644993 10 38 38 38 38 838876
25 37.2478948021 2.58021368225 10 38 38 38 38 838876
26 37.1056830807 3.04029966339 10 38 38 38 38 838876
27 37.0417129588 3.21755695865 9 38 38 38 38 838876
28 36.9245287742 3.46922882082 9 38 38 38 38 838876
29 37.0233538687 3.22132395112 9 38 38 38 38 838876
30 36.9768440151 3.36846192959 9 38 38 38 38 838876
31 36.9019378311 3.51733823479 10 38 38 38 38 838876
32 37.0442532627 3.15497307231 10 38 38 38 38 838876
33 37.0763462061 3.05842992907 10 38 38 38 38 838876
34 36.9112836701 3.51048896466 10 38 38 38 38 838876
35 36.871104907 3.50954115324 10 37 38 38 38 838876
36 36.9835911386 3.29022069717 9 38 38 38 38 838876
37 36.9526103977 3.40894509478 9 38 38 38 38 838876
38 36.9730198504 3.35198104312 10 38 38 38 38 838876
39 36.925962836 3.42925499742 9 38 38 38 38 838876
40 36.9641508399 3.3749237703 10 38 38 38 38 838876
41 36.9701290775 3.37108719426 9 38 38 38 38 838876
42 36.925310773 3.46541098262 9 38 38 38 38 838876
43 36.4323821399 4.37591782904 9 37 38 38 38 838876
44 36.6849510536 3.84804558398 10 37 38 38 38 838876
45 36.8000574578 3.71757395575 10 37 38 38 38 838876
46 36.743696327 3.89392229051 9 37 38 38 38 838876
47 36.6721195981 4.06597933483 10 37 38 38 38 838876
48 36.7998965282 3.73484635736 9 37 38 38 38 838876
49 36.9068074423 3.46537970313 10 38 38 38 38 838876
50 36.828634983 3.67970695764 10 37 38 38 38 838876
51 36.7344208202 3.75309659394 9 37 38 38 38 838876
Average quality string
BBBBBFFFFFFFFFFFFFFFFFEFFFFEFEEFFEEEEEEEEEEEEEEEEEE
| FILE SIZE: | 85M |
|---|---|
| TIME TAKEN: | 1.10m |
Basic analysis on Alignment files (BAM)¶
Example: Check bam headers
1 | samtools view -H /export/NFS/Saranga/BaseSpace/Projects/MiSeq\ v3\:\ TruSeq\ Targeted\ RNA\ Expression\ \(NFkB\ \&\ Cell\ Cycle\:\ Human\ Brain-Liver-UHRR\)/AppSessions/TopHat\ Alignment\:\ No\ cSNP\ or\ RNA\ Editing\ found\ \(36\ Samples\)/AppResults.26970091.Brain1/Files/alignments/Brain1.alignments.bam
|
@HD VN:1.0 SO:coordinate
@RG ID:19 SM:Brain1
@SQ SN:chrM LN:16571
@SQ SN:chr1 LN:249250621
@SQ SN:chr2 LN:243199373
@SQ SN:chr3 LN:198022430
@SQ SN:chr4 LN:191154276
@SQ SN:chr5 LN:180915260
@SQ SN:chr6 LN:171115067
@SQ SN:chr7 LN:159138663
@SQ SN:chr8 LN:146364022
@SQ SN:chr9 LN:141213431
@SQ SN:chr10 LN:135534747
@SQ SN:chr11 LN:135006516
@SQ SN:chr12 LN:133851895
@SQ SN:chr13 LN:115169878
@SQ SN:chr14 LN:107349540
@SQ SN:chr15 LN:102531392
@SQ SN:chr16 LN:90354753
@SQ SN:chr17 LN:81195210
@SQ SN:chr18 LN:78077248
@SQ SN:chr19 LN:59128983
@SQ SN:chr20 LN:63025520
@SQ SN:chr21 LN:48129895
@SQ SN:chr22 LN:51304566
@SQ SN:chrX LN:155270560
@SQ SN:chrY LN:59373566
@PG ID:TopHat VN:2.0.7 CL:/illumina/development/IsisRNA/2.4.19.5/IsisRNA_Tools/bin/tophat --bowtie1 --read-realign-edit-dist 1 --segment-length 24 -o /data/input/Alignment/samples/Brain1/replicates/Brain1/tophat_main -p 1 --GTF /illumina/development/Genomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf --rg-id 19 --rg-sample Brain1 --library-type fr-firststrand --no-coverage-search --keep-fasta-order /illumina/development/Genomes/Homo_sapiens/UCSC/hg19/Sequence/BowtieIndex/genome /data/input/Alignment/samples/Brain1/replicates/Brain1/filtered/Brain1_S20_L001_R1_001.fastq.gz
| FILE SIZE: | 17M |
|---|---|
| TIME TAKEN: | 0.006s |
Example: Check basic statistics on a Bam file
1 | bamutils stats /export/NFS/Saranga/BaseSpace/Projects/MiSeq\ v3\:\ TruSeq\ Targeted\ RNA\ Expression\ \(NFkB\ \&\ Cell\ Cycle\:\ Human\ Brain-Liver-UHRR\)/AppSessions/TopHat\ Alignment\:\ No\ cSNP\ or\ RNA\ Editing\ found\ \(36\ Samples\)/AppResults.26970091.Brain1/Files/alignments/Brain1.alignments.bam
|
Reads: 766531
Mapped: 766531
Unmapped: 0
Flag distribution
[0x010] Reverse complimented 383242 50.00%
[0x100] Secondary alignment 47 0.01%
Reference counts count
chr1 32252
chr10 6829
chr11 45127
chr12 74524
chr13 24336
chr14 81664
chr15 254
chr16 5704
chr17 46662
chr18 9922
chr19 25644
chr2 32527
chr20 10708
chr21 22
chr22 7805
chr3 83585
chr4 42487
chr5 47865
chr6 106512
chr7 18024
chr8 6921
chr9 25334
chrM 0
chrX 31823
chrY 0
Metadata¶
In each directory, BaseMount provides a number of hidden files with extra BaseSpace metadata. These are hidden files and their names start with a “.”.
1 2 | cd /export/NFS/Saranga/BaseSpace/Projects/MiSeq\ v3\:\ TruSeq\ Targeted\ RNA\ Expression\ \(NFkB\ \&\ Cell\ Cycle\:\ Human\ Brain-Liver-UHRR\)/Samples/
ls -l .?* #List only the hidden files
|
-r-------- 1 swijeratne swijeratne 149 Nov 16 11:29 .curl
.
.
.
-r--r--r-- 1 swijeratne swijeratne 40674 Nov 16 11:29 .json
Display content of the .json file
1 | cat .json
|
Query through a .json file qith “jq”
1 2 3 | cat .json | jq '.Response.Items[] | select(.NumReadsPF ) | {Name: .Name, PF : .NumReadsPF}'
cat .json | jq '.Response.Items[] | select(.NumReadsPF ) | "\( .Name)\t\(.NumReadsPF)"'
cat .json | jq '.Response.Items[] | select(.NumReadsPF > 747912) | "\( .Name)\t\(.NumReadsPF)"'
|
Limitations of BaseMount¶
According to Illumina,
Every new directory access made by BaseMount relies on FUSE, the BaseSpace API and the user’s credentials. This mechanism means that, as currently available, BaseMount does not support the following types of access:
- Cluster access, where many compute nodes can access the files. FUSE mounted file systems are per-host and cannot be accessed from many hosts without additional infrastructure.
- BaseMount also doesn’t refresh files or directories. In order to reflect changes done via the Web GUI in your command line tree, you currently need to unmount (basemount –unmount ) and restart BaseMount.
- The Runs Files directory is not mounted automatically for you as there can be 100k + files available in that mount which can take a couple minutes to load for really large runs. You can still mount this directory manually if needed.
- In general, lots of concurrent requests can cause stability issues on a resource constrained system. Keep in mind, this is an early release and stability will increase.